
I'm a data scientist at Molcure Inc. in Tokyo, Japan, with training in neuroscience. My primary interests lie in machine learning and protein modeling, particularly the use of natural language processing to decipher the hidden grammar of protein sequences.
My Work
Conference Papers
Dorfer TA, Robinson CN, Cocchi L, Mattingley JB, Sale MV, Zalesky A, Gollo LL. Whole-brain network states predict behavioral responses to transcranial magnetic stimulation. Imaging@Brisbane, Brisbane, August 2018.
Pater MRA, Dorfer TA, Gschwind L, Coynel D, Papassotiropoulos A, de Quervain DJ, Luksys G. Predicting Human Memory Performance through Multi-Voxel Pattern Analysis. Federation of European Neuroscience Societies, Berlin, July 2018.
Dorfer TA, Roberts JA, Breakspear M, Gollo LL. Slow oscillatory brain activity renders convolution with the hemodynamic response function redundant. Organization for Human Brain Mapping, Singapore, June 2018.
Dorfer TA, Pater M, Gschwind L, Papassotiropoulos A, de Quervain DJ, Luksys G. FMRI-based prediction models for free recall, recognition memory, emotional valences, arousal, and memorability of pictures. Organization for Human Brain Mapping, Singapore, June 2018.
Gollo LL, Cocchi L, Hearne L, Dorfer TA, Roberts J, Breakspear M. Can we predict the intensity of the effects of brain stimulation? Organization for Human Brain Mapping, Singapore, June 2018.
ProtLearn
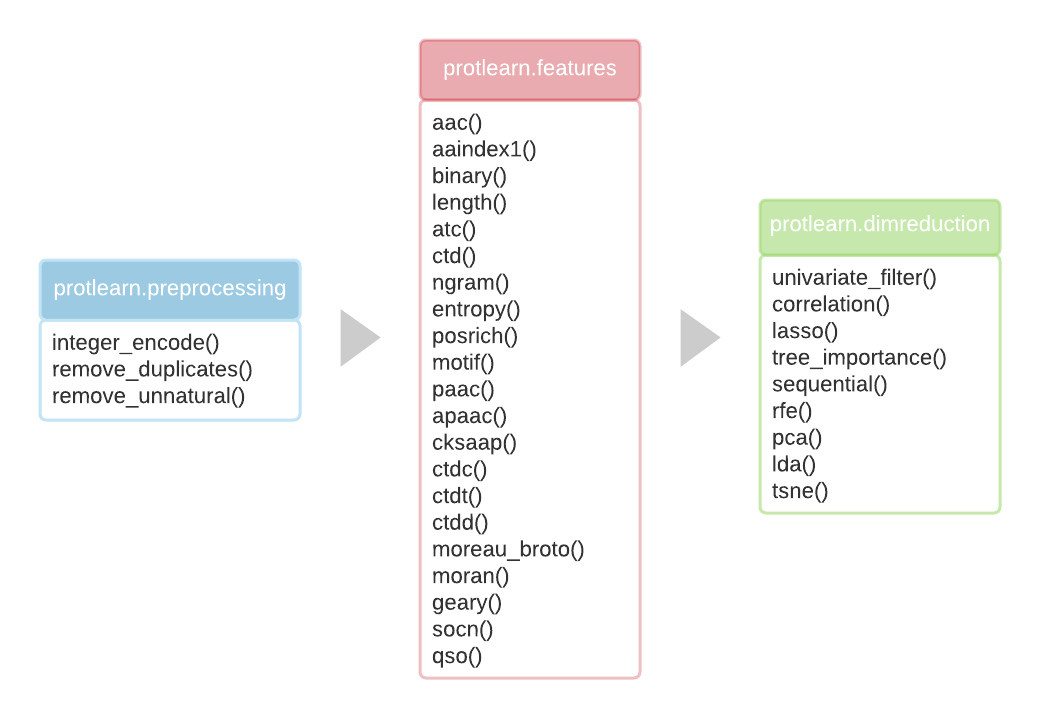
ProtLearn is a feature extraction tool for protein sequences. It is a freely available Python package that allows the user to efficiently extract amino acid sequence features from proteins and peptides, which can then be used for a variety of downstream machine learning tasks.
Natural Language Processing for Proteins (NLProt)
The application of Natural Language Processing (NLP) to protein sequence prediction has recently gained traction in the fields of machine learning and computational biology. This was primarily fueled by the recent advances in deep learning and language models such as Google's BERT and its successors RoBERTa and ALBERT. The aim of this site is to provide a comprehensive and chronologically ordered list of the recently published literature in this area. If you have come across relevant work that should be added to this list, please feel free to make a pull request or open an issue here.
Papers (blobs) arranged by similarity and date (the more recent, the bigger).
Technical Writing
Dynamic Replay of time-series data | Utilizing matplotlib and double-ended queues in Python
April 6, 2020 · 2min read · Read article in Towards Data Science
Artefact Correction with ICA | Illustrated with an example from the neurosciences
April 4, 2020 · 5min read · Read article in Towards Data Science
Dynamic Replay of time-series data | Utilizing matplotlib and double-ended queues in Python
April 6, 2020 · 2min read · Read article in Towards Data Science
Artefact Correction with ICA | Illustrated with an example from the neurosciences
April 4, 2020 · 5min read · Read article in Towards Data Science